Mike's NAO notebook
Explore DITP WastewaterSCAN data
Guide to estimating recovery efficiency using a spike-in
Some draft notes to support design and analysis for upcoming experiments.
Evaluate viral MGS protocols: Amicon centrifugal filters vs InnovaPrep Concentrating Pipette
Evaluate viral MGS protocols: Dissociation treatments and Amicon MWCOs
Reanalysis of Conceição-Neto et al 2015 protocol testing
WWTP basic detection theory
Notes exploring the factors that determine our ability to perform _basic detection_ in a WWTP setting. By basic detection, I mean detection based on seeing a sufficient number of distinguishing reads from the pathogen, rather than from a spatiotemporal pattern such as exponential growth.
Concentration tests
Tests of wastewater concentration methods performed by Ari in May 2023.
Analyze phagemid qPCR data for creating spike-in stock
Review the 2023-04 stock quantification experiment
Examining the effect of noise on Exponential Growth Detection
EGD theory notes
Captured notes on exponential growth detection.
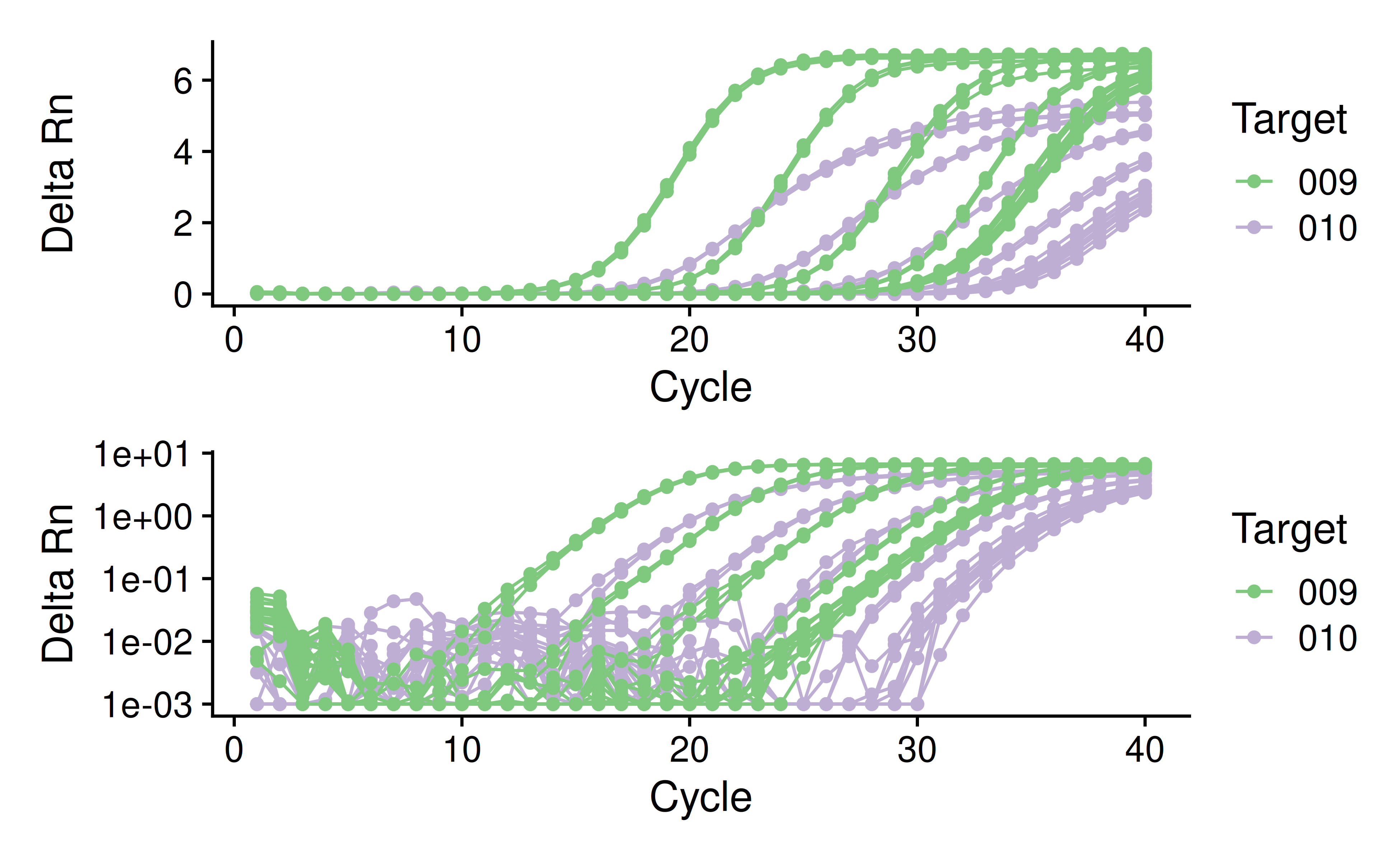
Developing a qPCR data analysis workflow
In-progress qPCR data analysis workflow in R, using data from the 2022-Q2 Sprint spike-in experiment.
CZID-to-R data import and analysis demo
Demonstration of how to import taxonomic profiles from CZID into R and do a few basic analyses.